Tuesday Poster Session
Category: IBD
P4267 - Identification of New Therapeutic Targets in IBD Using a Transcriptomic Foundation Model
Tuesday, October 29, 2024
10:30 AM - 4:00 PM ET
Location: Exhibit Hall E
Has Audio
- VB
Vincent Bouget, MSc
Scienta Lab
Paris, Ile-de-France, France
Presenting Author(s)
Award: Presidential Poster Award
Apolline Bruley, PhD, Vincent Bouget, MSc, Julien Duquesne, MSc
Scienta Lab, Paris, Ile-de-France, France
Introduction: Drug reponse in inflammatory bowel diseases (IBD) is heterogenous and faces a therapeutic ceiling, identifying new therapeutic targets is needed to improve IBD management.
This study is a large-scale reanalysis of multiple transcriptomics datasets to identify new therapeutic targets. A foundation model was used to compare RNA data in IBD and healthy samples.
Methods: We selected 59 public bulk RNAseq datasets from GEO and GTEx. We included RNA data from biopsies of the gastrointestinal (GI) tract of IBD patients and healthy controls (HC). Each dataset went through QA/QC, normalization, and binning to enable inter-datasets comparison. IBD datasets were then pooled (5,902 samples); as for HC datasets (1,317 samples).
To generate computational networks of IBD & HC transcriptomes, a deep learning RNA foundation model was separately finetuned on the IBD & HC datasets. Pairwise cosine similarities of gene embeddings were computed to create the network’s adjacency matrix. This resulted in 2 gene regulatory networks (IBD & HC), where each gene node is positively or negatively coregulated to its neighbors.
Gene’s connections were compared between HC & IBD networks, the 20 genes displaying the greatest differences with its neighbors were selected as potential therapeutic targets. Gene biological function was described with Reactome. A subnetwork with these 20 genes and licensed drug targets was computed.
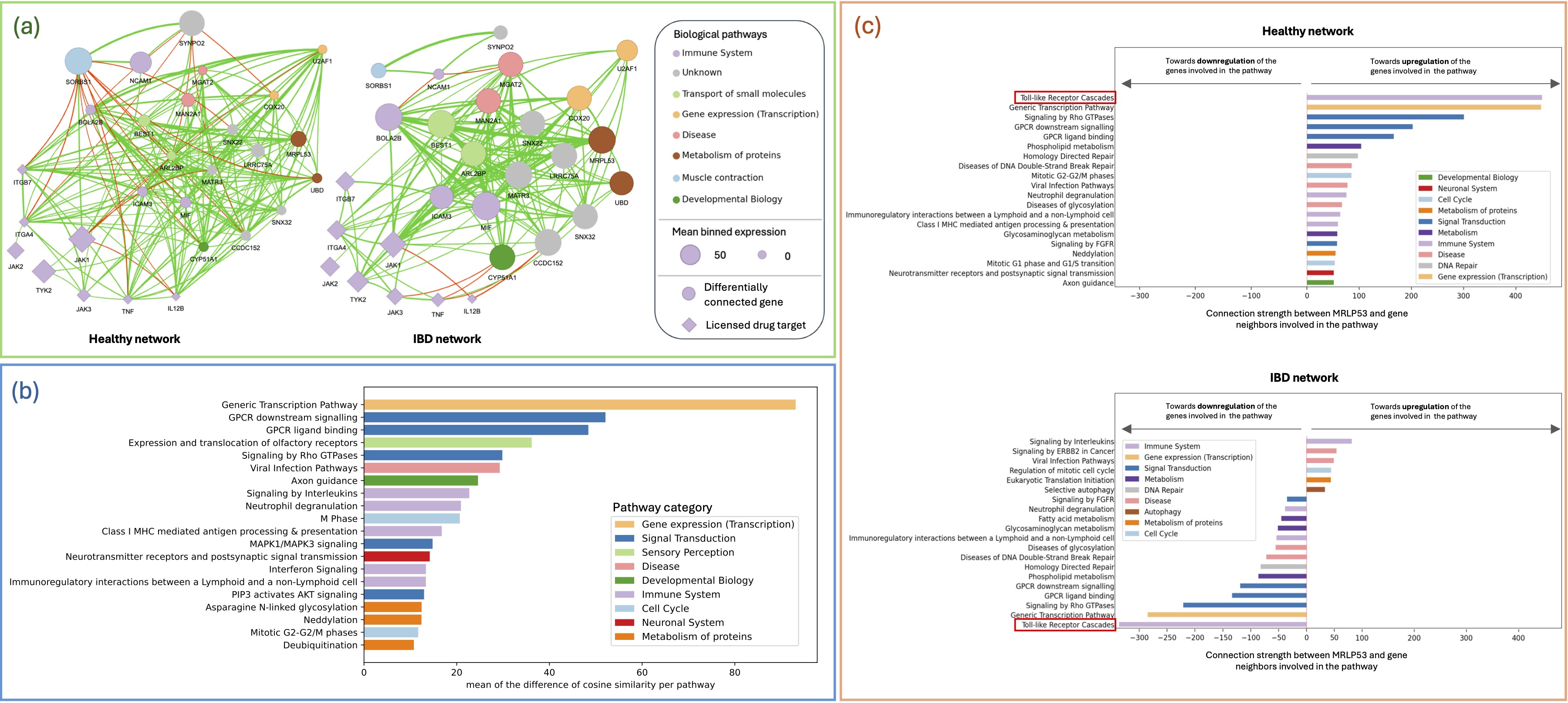
Results: The results are made available in an interactive webapp at enigma.scientalab.com. The subnetwork is depicted in Figure A. Interestingly, in addition to being differently connected, the 20 potential targets are also differentially expressed in IBD vs. HC: 17 are upregulated and 3 downregulated (p < 0.01). Of note, 10 are strongly connected to licensed drug targets; and 11 were identified as key modulators of IBDs (Table 1).
Biological pathways involving these 20 genes were differently activated in IBD vs. HC (Figure B), such as signaling by interleukins and neutrophil degranulation, 2 pathways of the immune system.
Discussion: The 20 genes identified are potential drug targets. For instance, MRPL53’s connections to genes involved in the immune system are activated in IBD and repressed in HC (Figure C). Of note, MRPL53's modifies toll-like receptor cascades in IBDs vs. HC, a pathway involved in bacterial infection response - consistent with aberrant immune responses to intestinal microorganisms in IBD. Next step is to prioritize these potential targets based on safety & druggability.
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Apolline Bruley, PhD, Vincent Bouget, MSc, Julien Duquesne, MSc. P4267 - Identification of New Therapeutic Targets in IBD Using a Transcriptomic Foundation Model, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
Apolline Bruley, PhD, Vincent Bouget, MSc, Julien Duquesne, MSc
Scienta Lab, Paris, Ile-de-France, France
Introduction: Drug reponse in inflammatory bowel diseases (IBD) is heterogenous and faces a therapeutic ceiling, identifying new therapeutic targets is needed to improve IBD management.
This study is a large-scale reanalysis of multiple transcriptomics datasets to identify new therapeutic targets. A foundation model was used to compare RNA data in IBD and healthy samples.
Methods: We selected 59 public bulk RNAseq datasets from GEO and GTEx. We included RNA data from biopsies of the gastrointestinal (GI) tract of IBD patients and healthy controls (HC). Each dataset went through QA/QC, normalization, and binning to enable inter-datasets comparison. IBD datasets were then pooled (5,902 samples); as for HC datasets (1,317 samples).
To generate computational networks of IBD & HC transcriptomes, a deep learning RNA foundation model was separately finetuned on the IBD & HC datasets. Pairwise cosine similarities of gene embeddings were computed to create the network’s adjacency matrix. This resulted in 2 gene regulatory networks (IBD & HC), where each gene node is positively or negatively coregulated to its neighbors.
Gene’s connections were compared between HC & IBD networks, the 20 genes displaying the greatest differences with its neighbors were selected as potential therapeutic targets. Gene biological function was described with Reactome. A subnetwork with these 20 genes and licensed drug targets was computed.
Results: The results are made available in an interactive webapp at enigma.scientalab.com. The subnetwork is depicted in Figure A. Interestingly, in addition to being differently connected, the 20 potential targets are also differentially expressed in IBD vs. HC: 17 are upregulated and 3 downregulated (p < 0.01). Of note, 10 are strongly connected to licensed drug targets; and 11 were identified as key modulators of IBDs (Table 1).
Biological pathways involving these 20 genes were differently activated in IBD vs. HC (Figure B), such as signaling by interleukins and neutrophil degranulation, 2 pathways of the immune system.
Discussion: The 20 genes identified are potential drug targets. For instance, MRPL53’s connections to genes involved in the immune system are activated in IBD and repressed in HC (Figure C). Of note, MRPL53's modifies toll-like receptor cascades in IBDs vs. HC, a pathway involved in bacterial infection response - consistent with aberrant immune responses to intestinal microorganisms in IBD. Next step is to prioritize these potential targets based on safety & druggability.

Figure: Figure 1. (A) Networks of the 20 most differentially connected genes between healthy and IBD network (circles) and of the targets of the commercialized IBD therapies (diamonds). Nodes represent the genes; their color represent the biological pathway they are involved in, and their size reflects their level of expression. Edges represent the relationships between the genes, a green edge symbolizes upregulation and a red edge downregulation. (B) Pathways most impacted by the top 20 differentially connected genes. For each gene, its connection with any gene involved in the same Reactome pathway is computed – this value is compared between IBD & HC networks, and averaged. (C) Pathways of the genes affected by the changes in the connections of MRPL53 in the healthy network (top) compared to the IBD network (bottom).
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Apolline Bruley indicated no relevant financial relationships.
Vincent Bouget indicated no relevant financial relationships.
Julien Duquesne indicated no relevant financial relationships.
Apolline Bruley, PhD, Vincent Bouget, MSc, Julien Duquesne, MSc. P4267 - Identification of New Therapeutic Targets in IBD Using a Transcriptomic Foundation Model, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.

