Sunday Poster Session
Category: Pediatrics
P1466 - Metagenomic Analysis of Gut Microbiota in Phenylketonuria (PKU) Patients: A New Perspective From the Ecuadorian Population
Sunday, October 27, 2024
3:30 PM - 7:00 PM ET
Location: Exhibit Hall E
.jpg)
Ariel L. Vargas, MD
Universidad San Francisco de Quito
Philadelphia, PA
Presenting Author(s)
Award: Presidential Poster Award
Ariel L. Vargas, MD1, Ivonne Salinas, MD2, Vanessa Romero, MD, PhD2, Paul Leon-Gomez, 2, Camila Gallegos, MD2
1Universidad San Francisco de Quito, Philadelphia, PA; 2Universidad San Francisco de Quito, Quito, Pichincha, Ecuador
Introduction: PKU is a disorder caused by a phenylalanine hydroxylase deficiency, essential for converting phenylalanine to tyrosine. If untreated, it leads to severe cognitive impairments. This study uses metagenomic sequencing to analyze the gut microbiome of PKU patients. PKU diet can influence on microbiome composition and an altered microbiome can, in turn, impact the efficacy of therapy and overall health.
Methods: After obtaining approval from the bioethics committee and written consent, we collected stool samples from all patients who met our criteria: diagnosis of PKU, age 2 and < 18 years, and no antibiotic use within the past 30 days. We performed DNA extraction using Qiagen kits, library preparation with Ilumina Nextera, and sequencing with Ilumina MiSeq v3. After data processing and quality control, we performed taxonomic annotation with Krona. Using Phyloseq, we obtained the final metagenomics data.
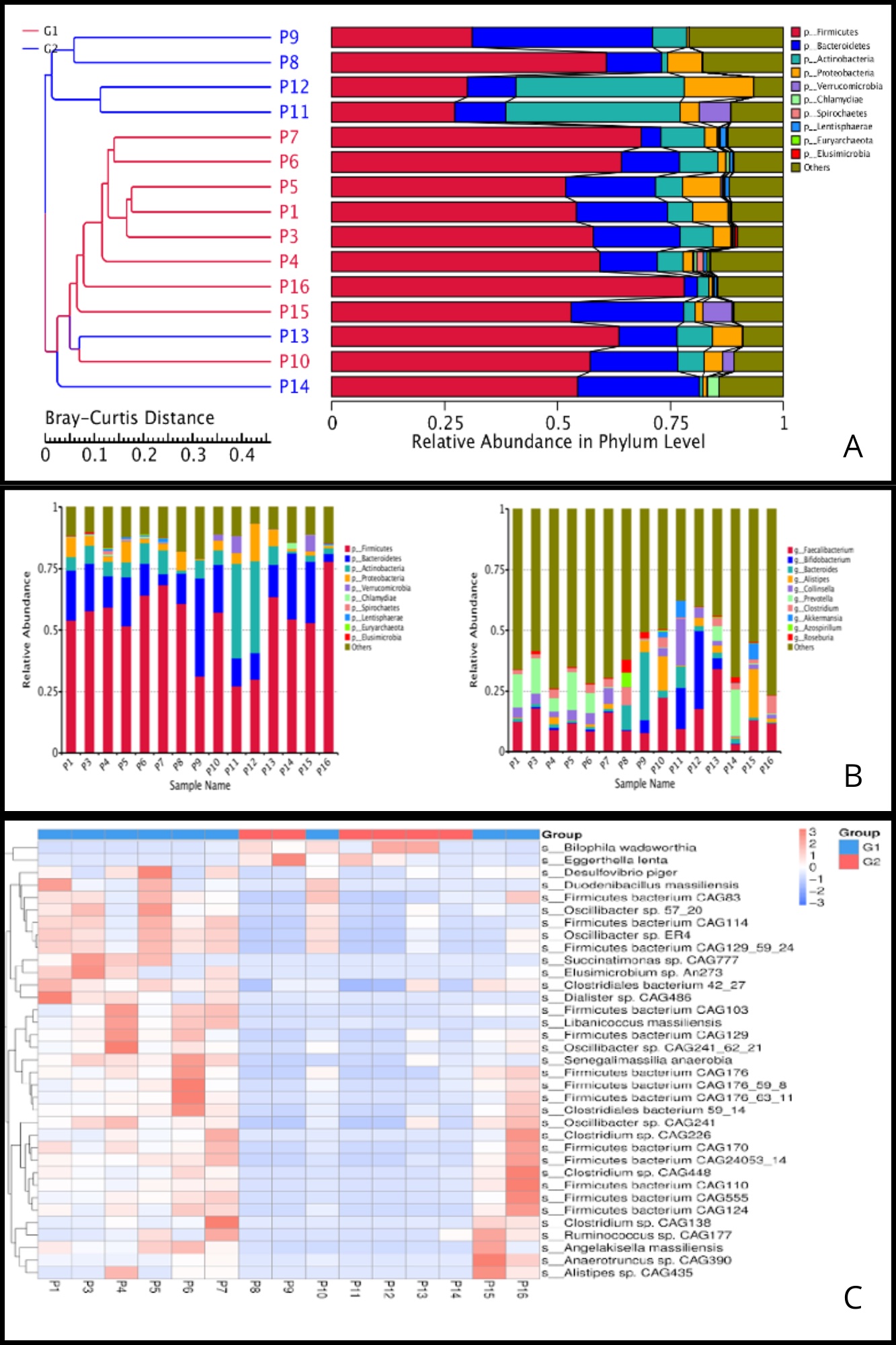
Results: In total, 15 patients from various regions of Ecuador were included: 9 on a liberalized diet and 6 on a Phe-restricted diet. The gut microbiota composition was analyzed and presented as relative abundance at phylum (Fig 1A) and genus levels (Fig 1B). Specific group differences for species are illustrated in Fig 1C.
Discussion: All food components are needed for nutrition and to serve as substrate for gut microbiota, which is increasingly recognized for its role in maintaining health. Phe-restricted diet can impact development and diversification of patients’ microbiomes.
G1 exhibited a gut microbiome somewhat similar to that of pediatric healthy patients, except for a less diverse microbiome, with enrichment in Firmicutes and Bacteroidetes, possibly due to the fact that their diet is unrestricted and aligns with that of the general population. Butyrate-producing genera are depleted in G2 due to the changes in available substrates caused by a high carbohydrate/fiber and low protein diet, leading to absence of their anti-inflammatory properties. Both groups are depleted in species that are considered potential probiotics due to their anti-inflammatory effects and role in maintaining gut homeostasis, such as Akkermansia.
Diet plays a pivotal role in regulating the microbiome, and changes in its composition can potentially affect microbiome architecture even among patients with the same condition. Further research is needed to uncover how changes in gut microbiome may impact response to therapy, disease control, susceptibility to other illnesses, and potential pathways for treatment.
Disclosures:
Ariel L. Vargas, MD1, Ivonne Salinas, MD2, Vanessa Romero, MD, PhD2, Paul Leon-Gomez, 2, Camila Gallegos, MD2. P1466 - Metagenomic Analysis of Gut Microbiota in Phenylketonuria (PKU) Patients: A New Perspective From the Ecuadorian Population, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
Ariel L. Vargas, MD1, Ivonne Salinas, MD2, Vanessa Romero, MD, PhD2, Paul Leon-Gomez, 2, Camila Gallegos, MD2
1Universidad San Francisco de Quito, Philadelphia, PA; 2Universidad San Francisco de Quito, Quito, Pichincha, Ecuador
Introduction: PKU is a disorder caused by a phenylalanine hydroxylase deficiency, essential for converting phenylalanine to tyrosine. If untreated, it leads to severe cognitive impairments. This study uses metagenomic sequencing to analyze the gut microbiome of PKU patients. PKU diet can influence on microbiome composition and an altered microbiome can, in turn, impact the efficacy of therapy and overall health.
Methods: After obtaining approval from the bioethics committee and written consent, we collected stool samples from all patients who met our criteria: diagnosis of PKU, age 2 and < 18 years, and no antibiotic use within the past 30 days. We performed DNA extraction using Qiagen kits, library preparation with Ilumina Nextera, and sequencing with Ilumina MiSeq v3. After data processing and quality control, we performed taxonomic annotation with Krona. Using Phyloseq, we obtained the final metagenomics data.
Results: In total, 15 patients from various regions of Ecuador were included: 9 on a liberalized diet and 6 on a Phe-restricted diet. The gut microbiota composition was analyzed and presented as relative abundance at phylum (Fig 1A) and genus levels (Fig 1B). Specific group differences for species are illustrated in Fig 1C.
Discussion: All food components are needed for nutrition and to serve as substrate for gut microbiota, which is increasingly recognized for its role in maintaining health. Phe-restricted diet can impact development and diversification of patients’ microbiomes.
G1 exhibited a gut microbiome somewhat similar to that of pediatric healthy patients, except for a less diverse microbiome, with enrichment in Firmicutes and Bacteroidetes, possibly due to the fact that their diet is unrestricted and aligns with that of the general population. Butyrate-producing genera are depleted in G2 due to the changes in available substrates caused by a high carbohydrate/fiber and low protein diet, leading to absence of their anti-inflammatory properties. Both groups are depleted in species that are considered potential probiotics due to their anti-inflammatory effects and role in maintaining gut homeostasis, such as Akkermansia.
Diet plays a pivotal role in regulating the microbiome, and changes in its composition can potentially affect microbiome architecture even among patients with the same condition. Further research is needed to uncover how changes in gut microbiome may impact response to therapy, disease control, susceptibility to other illnesses, and potential pathways for treatment.

Figure: Figure 1. A. Sample clustering tree with taxonomic abundance at phylum level. PKU patients were segregated into groups based on their nutritional therapy status: those on a liberalized or non-Phe-restricted diet (G1) and those managed with a Phe-restricted diet (G2). G1 patients were notably enriched in Firmicutes and, to a lesser extent, Bacteroidetes, while showing depletion in Actinobacteria, Proteobacteria, and Verrucomicrobia. G2 patients exhibited various results: P8, P13, and P14 showed enrichment in Firmicutes but depletion in Bacteroidetes and other phyla. Conversely, P9, P11, and P12 had higher levels of Bacteroidetes and Actinobacteria, with lower quantities of Firmicutes. B. Relative abundance at the genus level. For both groups, the ‘others’ group represents the majority of the genera identified, suggesting a potential microbiome disruption in the sample processing phase. Besides this finding, G1 patients were enriched in Faecalibacterium and Prevotella genera, except for P10 and P15 who also had abundant Alistipes genus, and P16, who had Clostridium enrichment. G2 patients had higher levels of Bacteroides and Bifidobacterium, with a modest increase in Prevotella and Firmicutes among a few patients within the group, specifically P14 and P13, respectively. There was depletion of the Akkermansia and Roseburia genera across all patients from both groups. C. Abundance heatmap based on significantly variant species. X-axis represents the sample information, and the Y-axis represents species annotation; the left side of the heatmap shows the clustering tree of species. G1 patients have a significant predominance of Firmicutes, Oscillibacter, and Clostridium species, whereas G2 patients have a microbiome depleted in these species and abundant in Bilophila wadsworthia and Eggerthella lenta.
Disclosures:
Ariel Vargas indicated no relevant financial relationships.
Ivonne Salinas indicated no relevant financial relationships.
Vanessa Romero indicated no relevant financial relationships.
Paul Leon-Gomez indicated no relevant financial relationships.
Camila Gallegos indicated no relevant financial relationships.
Ariel L. Vargas, MD1, Ivonne Salinas, MD2, Vanessa Romero, MD, PhD2, Paul Leon-Gomez, 2, Camila Gallegos, MD2. P1466 - Metagenomic Analysis of Gut Microbiota in Phenylketonuria (PKU) Patients: A New Perspective From the Ecuadorian Population, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.

