Monday Poster Session
Category: Liver
P2976 - Deciphering Survival Disparities in Hepatocellular Carcinoma: A Multi-Omics Approach
Monday, October 28, 2024
10:30 AM - 4:00 PM ET
Location: Exhibit Hall E
Has Audio

Basile Njei, MD
Yale University School of Medicine
New Haven, CT
Presenting Author(s)
Award: Presidential Poster Award
Lydia Fozo, 1, Leila Ghaffari, BS2, Yazan Al-Ajlouni, MD3, Basile Njei, MD4
1Harvard University, Cambridge, MA; 2University of Maryland, Baltimore, MD; 3Staten Island University Hospital, Northwell Health, New York, NY; 4Yale University School of Medicine, New Haven, CT
Introduction: Hepatocellular carcinoma (HCC) exhibits significant survival disparities driven by its complex molecular and microbial landscape. This study aims to elucidate the prognostic markers and underlying biological pathways contributing to these disparities by integrating genomic, proteomic, epigenomic, and microbiome profiling.
Methods: We conducted a multi-omics analysis using data from The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection (TCGA-LIHC) and Gene Expression Omnibus (GSE14520) for training, and the International Cancer Genome Consortium (ICGC-LIRI-JP) for external validation (Table 1). Cox regression and LASSO regression risk modeling were employed to assess genes, proteins, DNA methylation, and microbiome signatures associated with poor survival, using Bonferroni correction of P-values.
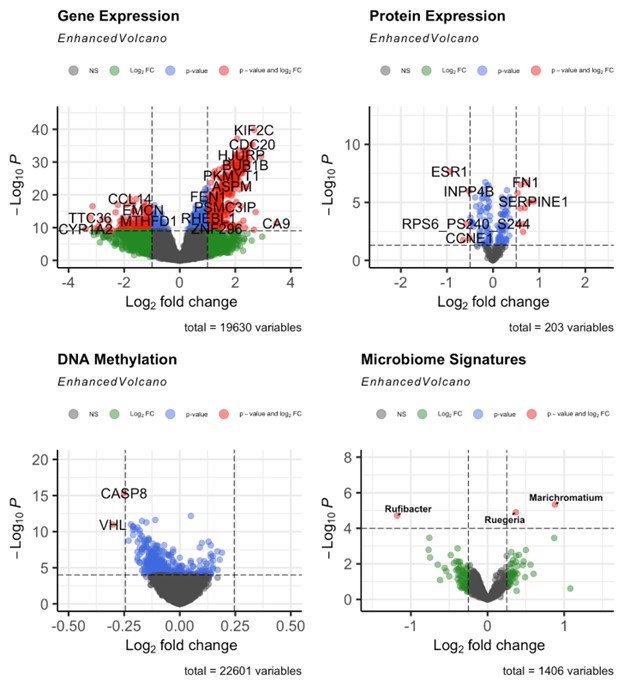
Results: Our integrated analysis identified 24 common prognostic markers (Figure 1) across genomic, proteomic, and epigenomic datasets, with seven markers (EXO1, KIF20A, MCM2, PKM, SFN, IGFALS, and RHOBTB3) selected for the LASSO-based risk model. This model achieved a 1-year and 5-year survival prediction area under the curve (AUC) of 0.82 and 0.70 respectively, in the validation subset. Gene set enrichment analysis revealed that high-risk groups exhibited upregulation in immune regulatory pathways, while low-risk groups showed enrichment in metabolic and signaling pathways. Epigenomic analysis highlighted increased methylation of VHL and CASP8 in high-risk groups, potentially promoting cancer progression. Microbiome profiling revealed distinct microbial signatures between high and low-risk groups, suggesting their role in modulating HCC progression and patient survival, such as Rufibacter, which belongs to the Faecalibacterium family, a class of butyrate producers with anti-tumor properties.
Discussion: Integrating genomic, proteomic, epigenomic, and microbiome profiling provides a comprehensive understanding of survival disparities in HCC. Our study identifies critical molecular and microbial signatures associated with patient outcomes and develops a robust multi-omics risk model for clinical application. These findings underscore the importance of a multi-omics approach in uncovering the complex interplay between various biological factors in HCC. Future research should focus on validating these findings in larger cohorts and exploring their potential for guiding personalized therapeutic strategies to enhance survival and quality of life for HCC patients.
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Lydia Fozo, 1, Leila Ghaffari, BS2, Yazan Al-Ajlouni, MD3, Basile Njei, MD4. P2976 - Deciphering Survival Disparities in Hepatocellular Carcinoma: A Multi-Omics Approach, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
Lydia Fozo, 1, Leila Ghaffari, BS2, Yazan Al-Ajlouni, MD3, Basile Njei, MD4
1Harvard University, Cambridge, MA; 2University of Maryland, Baltimore, MD; 3Staten Island University Hospital, Northwell Health, New York, NY; 4Yale University School of Medicine, New Haven, CT
Introduction: Hepatocellular carcinoma (HCC) exhibits significant survival disparities driven by its complex molecular and microbial landscape. This study aims to elucidate the prognostic markers and underlying biological pathways contributing to these disparities by integrating genomic, proteomic, epigenomic, and microbiome profiling.
Methods: We conducted a multi-omics analysis using data from The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection (TCGA-LIHC) and Gene Expression Omnibus (GSE14520) for training, and the International Cancer Genome Consortium (ICGC-LIRI-JP) for external validation (Table 1). Cox regression and LASSO regression risk modeling were employed to assess genes, proteins, DNA methylation, and microbiome signatures associated with poor survival, using Bonferroni correction of P-values.
Results: Our integrated analysis identified 24 common prognostic markers (Figure 1) across genomic, proteomic, and epigenomic datasets, with seven markers (EXO1, KIF20A, MCM2, PKM, SFN, IGFALS, and RHOBTB3) selected for the LASSO-based risk model. This model achieved a 1-year and 5-year survival prediction area under the curve (AUC) of 0.82 and 0.70 respectively, in the validation subset. Gene set enrichment analysis revealed that high-risk groups exhibited upregulation in immune regulatory pathways, while low-risk groups showed enrichment in metabolic and signaling pathways. Epigenomic analysis highlighted increased methylation of VHL and CASP8 in high-risk groups, potentially promoting cancer progression. Microbiome profiling revealed distinct microbial signatures between high and low-risk groups, suggesting their role in modulating HCC progression and patient survival, such as Rufibacter, which belongs to the Faecalibacterium family, a class of butyrate producers with anti-tumor properties.
Discussion: Integrating genomic, proteomic, epigenomic, and microbiome profiling provides a comprehensive understanding of survival disparities in HCC. Our study identifies critical molecular and microbial signatures associated with patient outcomes and develops a robust multi-omics risk model for clinical application. These findings underscore the importance of a multi-omics approach in uncovering the complex interplay between various biological factors in HCC. Future research should focus on validating these findings in larger cohorts and exploring their potential for guiding personalized therapeutic strategies to enhance survival and quality of life for HCC patients.

Figure: Volcano plot depicting differential profiles between risk groups, with the low-risk group as the reference ( log2FC>0 is upregulated in high risk,log2FC<0 is downregulated in high risk) and the y-axis indicating statistical significance (-log10 adjusted p-value).
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Lydia Fozo indicated no relevant financial relationships.
Leila Ghaffari indicated no relevant financial relationships.
Yazan Al-Ajlouni indicated no relevant financial relationships.
Basile Njei indicated no relevant financial relationships.
Lydia Fozo, 1, Leila Ghaffari, BS2, Yazan Al-Ajlouni, MD3, Basile Njei, MD4. P2976 - Deciphering Survival Disparities in Hepatocellular Carcinoma: A Multi-Omics Approach, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.

