Monday Poster Session
Category: Liver
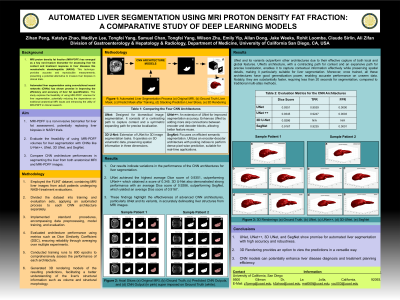
P2853 - Automated Liver Segmentation Using MRI Proton Density Fat Fraction: A Comparative Study of Deep Learning Models
Monday, October 28, 2024
10:30 AM - 4:00 PM ET
Location: Exhibit Hall E
Has Audio
- AZ
Ali Zifan, PhD
UC San Diego
La Jolla, CA
Presenting Author(s)
Ali Zifan, PhD1, Zihan Peng, BSc1, Katelyn Zhao, 1, Madilyn Lee, 1, Tongfei Yang, 1, Samuel Chan, 1, Wilson Zhu, 1, Emily Yip, 1, Allan Dong, 1, Jake Weeks, BS1, Rohit Loomba, MD, MHSc2, Claude Sirlin, MD1
1UC San Diego, La Jolla, CA; 2University of California San Diego School of Medicine, La Jolla, CA
Introduction: : Magnetic resonance imaging proton density fat fraction (MRI-PDFF) is a quantitative biomarker enabling accurate, repeatable, and reproducible assessment of liver fat across its entirety. As a non-invasive tool, MRI-PDFF is increasingly recognized as a leading surrogate to liver biopsy for evaluating treatment response in nonalcoholic steatohepatitis (NASH) trials. This study aimed to investigate the feasibility of using MRI-PDFF volumes directly for automated liver segmentation, potentially eliminating the need for traditional anatomical MRI inputs.
Methods: Data from 227 patients, including 114 from the FLINT study (mean age: 51.2 years, SD: 11.5; mean BMI: 33.6, SD: 5.14) and 113 from the CyNCh study (mean age: 13.7 years, SD: 11.5; mean BMI: 32, SD: 6.8), were utilized. Anatomical images were sourced from the 5th-echo of a LipoQuant 6-echo gradient-recalled echo (GRE) sequence, selected for its high contrast among the phased echoes. PDFF maps, derived from these echoes, were computed pixel-by-pixel using software on the scanner, employing nonlinear least-squares fitting or an Osirix/Horos image processing plugin. Advanced convolutional neural network (CNN) models, including U-Net++ and SegNet, were optimized and trained using radiologist-annotated masks derived from MRI images. Two mappings were examined: 1) direct mapping from anatomical images to whole liver segmentation, and 2) direct mapping from PDFF volumes to whole liver segmentation. Both models underwent optimization and training over 800 epochs on the combined dataset.
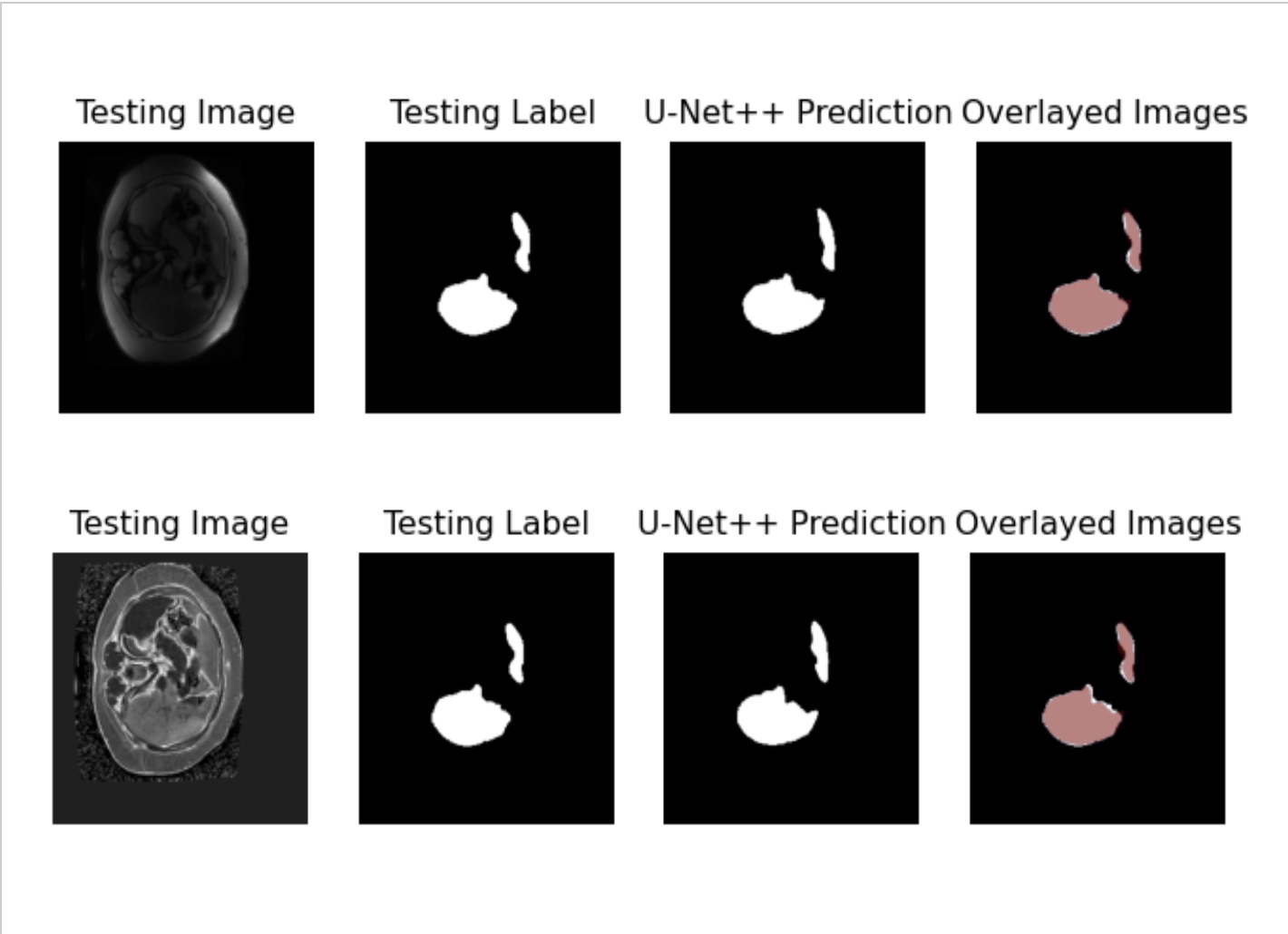
Results: U-Net++ achieved a Dice Score of 0.9409, True Positive Rate (TPR) of 0.9322, and False Positive Rate (FPR) of 0.0003 for anatomical images and whole liver segmentation. SegNet demonstrated similar performance with a Dice Score of 0.9356, TPR of 0.9364, and FPR of 6.91 x 10^-5. For MRI-PDFF volumes and whole liver segmentation, U-Net++ achieved a Dice Score of 0.9345, TPR of 0.9247, and FPR of 0.0003, while SegNet showed slightly lower results with a Dice Score of 0.9167, TPR of 0.9235, and FPR of 0.0001.
Discussion: Both U-Net++ and SegNet exhibited high performance in liver segmentation using both anatomical and MRI-PDFF images. These findings indicate the potential of MRI-PDFF as a viable alternative to anatomical MRI in clinical trials, potentially reducing the reliance on invasive liver biopsies and enhancing the efficiency of liver fat quantification in liver disease research.
Disclosures:
Ali Zifan, PhD1, Zihan Peng, BSc1, Katelyn Zhao, 1, Madilyn Lee, 1, Tongfei Yang, 1, Samuel Chan, 1, Wilson Zhu, 1, Emily Yip, 1, Allan Dong, 1, Jake Weeks, BS1, Rohit Loomba, MD, MHSc2, Claude Sirlin, MD1. P2853 - Automated Liver Segmentation Using MRI Proton Density Fat Fraction: A Comparative Study of Deep Learning Models, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
1UC San Diego, La Jolla, CA; 2University of California San Diego School of Medicine, La Jolla, CA
Introduction: : Magnetic resonance imaging proton density fat fraction (MRI-PDFF) is a quantitative biomarker enabling accurate, repeatable, and reproducible assessment of liver fat across its entirety. As a non-invasive tool, MRI-PDFF is increasingly recognized as a leading surrogate to liver biopsy for evaluating treatment response in nonalcoholic steatohepatitis (NASH) trials. This study aimed to investigate the feasibility of using MRI-PDFF volumes directly for automated liver segmentation, potentially eliminating the need for traditional anatomical MRI inputs.
Methods: Data from 227 patients, including 114 from the FLINT study (mean age: 51.2 years, SD: 11.5; mean BMI: 33.6, SD: 5.14) and 113 from the CyNCh study (mean age: 13.7 years, SD: 11.5; mean BMI: 32, SD: 6.8), were utilized. Anatomical images were sourced from the 5th-echo of a LipoQuant 6-echo gradient-recalled echo (GRE) sequence, selected for its high contrast among the phased echoes. PDFF maps, derived from these echoes, were computed pixel-by-pixel using software on the scanner, employing nonlinear least-squares fitting or an Osirix/Horos image processing plugin. Advanced convolutional neural network (CNN) models, including U-Net++ and SegNet, were optimized and trained using radiologist-annotated masks derived from MRI images. Two mappings were examined: 1) direct mapping from anatomical images to whole liver segmentation, and 2) direct mapping from PDFF volumes to whole liver segmentation. Both models underwent optimization and training over 800 epochs on the combined dataset.
Results: U-Net++ achieved a Dice Score of 0.9409, True Positive Rate (TPR) of 0.9322, and False Positive Rate (FPR) of 0.0003 for anatomical images and whole liver segmentation. SegNet demonstrated similar performance with a Dice Score of 0.9356, TPR of 0.9364, and FPR of 6.91 x 10^-5. For MRI-PDFF volumes and whole liver segmentation, U-Net++ achieved a Dice Score of 0.9345, TPR of 0.9247, and FPR of 0.0003, while SegNet showed slightly lower results with a Dice Score of 0.9167, TPR of 0.9235, and FPR of 0.0001.
Discussion: Both U-Net++ and SegNet exhibited high performance in liver segmentation using both anatomical and MRI-PDFF images. These findings indicate the potential of MRI-PDFF as a viable alternative to anatomical MRI in clinical trials, potentially reducing the reliance on invasive liver biopsies and enhancing the efficiency of liver fat quantification in liver disease research.

Figure: Figure 1. Whole liver segmentation using PDFF volumes.
Disclosures:
Ali Zifan indicated no relevant financial relationships.
Zihan Peng indicated no relevant financial relationships.
Katelyn Zhao indicated no relevant financial relationships.
Madilyn Lee indicated no relevant financial relationships.
Tongfei Yang indicated no relevant financial relationships.
Samuel Chan indicated no relevant financial relationships.
Wilson Zhu indicated no relevant financial relationships.
Emily Yip indicated no relevant financial relationships.
Allan Dong indicated no relevant financial relationships.
Jake Weeks indicated no relevant financial relationships.
Rohit Loomba: 89BIO – Consultant. Aardvark – Consultant. Altimmune – Consultant. Madrigal Pharmaceuticals – Consultant. Merck – Consultant. Novo Nordisk – Consultant. Takeda – Consultant. Terns – Consultant. Viking – Consultant.
Claude Sirlin indicated no relevant financial relationships.
Ali Zifan, PhD1, Zihan Peng, BSc1, Katelyn Zhao, 1, Madilyn Lee, 1, Tongfei Yang, 1, Samuel Chan, 1, Wilson Zhu, 1, Emily Yip, 1, Allan Dong, 1, Jake Weeks, BS1, Rohit Loomba, MD, MHSc2, Claude Sirlin, MD1. P2853 - Automated Liver Segmentation Using MRI Proton Density Fat Fraction: A Comparative Study of Deep Learning Models, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
