Sunday Poster Session
Category: Colon
P0202 - Antimicrobial Resistance in Clostridioides difficile: Is It on the Rise?
Sunday, October 27, 2024
3:30 PM - 7:00 PM ET
Location: Exhibit Hall E

Phi Tran, DO
Baylor Scott & White Medical Center
Temple, TX
Presenting Author(s)
Phi Tran, DO1, Collin Telchik, MD1, Munok Hwang, MS2, Hosoon Choi, PhD2, Piyali Chatterjee, PhD2, Philip Toltzis, MD3, Curtis Donskey, MD4, Jennifer Cadnum, MS3, Christine Marlow, MS4, Chetan Jinadatha, MD2
1Baylor Scott & White Medical Center, Temple, TX; 2Olin E. Teague Veterans' Medical Center, Temple, TX; 3Rainbow Babies and Children's Hospital, Cleveland, OH; 4Louis Stokes Cleveland Veterans' Affairs Medical Center, Cleveland, OH
Introduction: Clostridioides difficile (C. diff) is an urgent antibiotic resistance threat according to Centers for Disease Control and Prevention (CDC). However, testing for antimicrobial resistance in C. diff is not included in current C. difficile infection (CDI) diagnostic algorithms. Although resistance to antibiotics used to treat CDI has been uncommon (1), there have been some reports of reduced susceptibility to fidaxomicin and vancomycin, and emergence of resistance to non-CDI antibiotics has played a major role in CDI outbreaks. Whole genome sequencing (WGS) can be an alternate way tto identify C. diff resistance trends among local isolates. In this study, we evaluated the use of WGS to identify C. diff resistance trends.
Methods: For C. diff isolates collected from 129 asymptomatically colonized infants and 74 adults with CDI in 2020-2022, WGS was performed using the NextSeq (Illumina Inc., CA) platform. The sequencing data was analyzed using ResFinder, a publicly available database of resistance genes, including to vancomycin (vanA, vanH, vanM, vanX, vanC, vanE, vanG, vanN, and vanL) and metronidazole (nimA, nimB, nimC, nimD, nimE, nimF, nimH, nimI, and nimJ). The output was organized to visualize the resistance genes prevalence among our isolates.
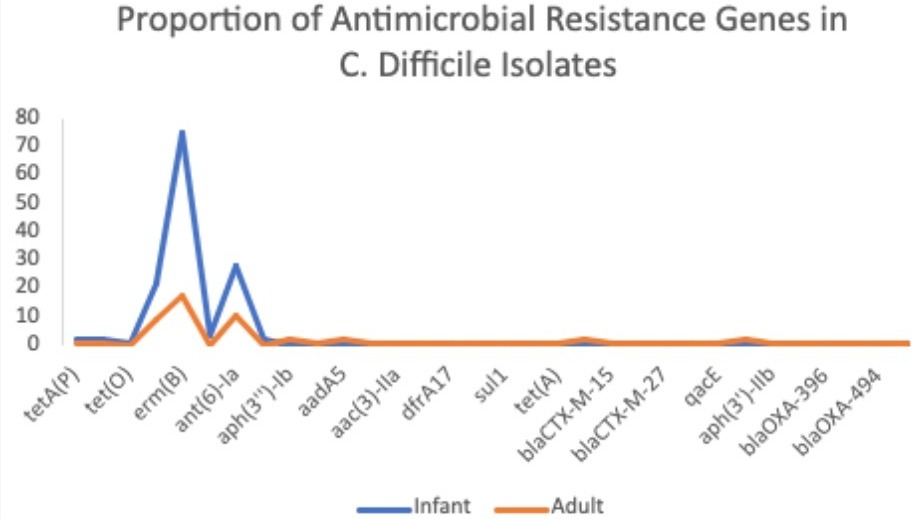
Results: We identified no vancomycin resistance genes amongst the 203 C. diff isolates. The most common resistance genes detected in both adult and pediatric isolates were the genetic mutation markers erm(B), ant(6)-la, and tet(M) which encode resistance to the non-CDI antibiotics clindamycin, streptomycin B, and tetracyclines, respectively (Figure 1).
Discussion: Our results suggest that WGS could be a useful tool to understand the prevalence of resistance genes in colonized or infected patients and to track the emergence of resistance. There is a need for the addition of newly identified genes encoding reduced susceptibility to fidaxomicin. Current treatment options for CDI are becoming limited due to global rising resistance. While ResFinder does not have all of the newly identified genes for Fidaxomicin in the database, many vancomycin and metronidazole resistance genes are present. We believe with updated ResFinder databases we can easily track the resistance of C. diff locally as well as in the United States to combat increasing resistance in an organism identified as an emergent threat.
Disclosures:
Phi Tran, DO1, Collin Telchik, MD1, Munok Hwang, MS2, Hosoon Choi, PhD2, Piyali Chatterjee, PhD2, Philip Toltzis, MD3, Curtis Donskey, MD4, Jennifer Cadnum, MS3, Christine Marlow, MS4, Chetan Jinadatha, MD2. P0202 - Antimicrobial Resistance in <i>Clostridioides difficile</i>: Is It on the Rise?, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
1Baylor Scott & White Medical Center, Temple, TX; 2Olin E. Teague Veterans' Medical Center, Temple, TX; 3Rainbow Babies and Children's Hospital, Cleveland, OH; 4Louis Stokes Cleveland Veterans' Affairs Medical Center, Cleveland, OH
Introduction: Clostridioides difficile (C. diff) is an urgent antibiotic resistance threat according to Centers for Disease Control and Prevention (CDC). However, testing for antimicrobial resistance in C. diff is not included in current C. difficile infection (CDI) diagnostic algorithms. Although resistance to antibiotics used to treat CDI has been uncommon (1), there have been some reports of reduced susceptibility to fidaxomicin and vancomycin, and emergence of resistance to non-CDI antibiotics has played a major role in CDI outbreaks. Whole genome sequencing (WGS) can be an alternate way tto identify C. diff resistance trends among local isolates. In this study, we evaluated the use of WGS to identify C. diff resistance trends.
Methods: For C. diff isolates collected from 129 asymptomatically colonized infants and 74 adults with CDI in 2020-2022, WGS was performed using the NextSeq (Illumina Inc., CA) platform. The sequencing data was analyzed using ResFinder, a publicly available database of resistance genes, including to vancomycin (vanA, vanH, vanM, vanX, vanC, vanE, vanG, vanN, and vanL) and metronidazole (nimA, nimB, nimC, nimD, nimE, nimF, nimH, nimI, and nimJ). The output was organized to visualize the resistance genes prevalence among our isolates.
Results: We identified no vancomycin resistance genes amongst the 203 C. diff isolates. The most common resistance genes detected in both adult and pediatric isolates were the genetic mutation markers erm(B), ant(6)-la, and tet(M) which encode resistance to the non-CDI antibiotics clindamycin, streptomycin B, and tetracyclines, respectively (Figure 1).
Discussion: Our results suggest that WGS could be a useful tool to understand the prevalence of resistance genes in colonized or infected patients and to track the emergence of resistance. There is a need for the addition of newly identified genes encoding reduced susceptibility to fidaxomicin. Current treatment options for CDI are becoming limited due to global rising resistance. While ResFinder does not have all of the newly identified genes for Fidaxomicin in the database, many vancomycin and metronidazole resistance genes are present. We believe with updated ResFinder databases we can easily track the resistance of C. diff locally as well as in the United States to combat increasing resistance in an organism identified as an emergent threat.

Figure: Figure 1: Proportion of Antimicrobial Resistance Gene Prevalence Among the 203 Clostridioides difficile isolates.
Disclosures:
Phi Tran indicated no relevant financial relationships.
Collin Telchik indicated no relevant financial relationships.
Munok Hwang indicated no relevant financial relationships.
Hosoon Choi indicated no relevant financial relationships.
Piyali Chatterjee indicated no relevant financial relationships.
Philip Toltzis indicated no relevant financial relationships.
Curtis Donskey indicated no relevant financial relationships.
Jennifer Cadnum indicated no relevant financial relationships.
Christine Marlow indicated no relevant financial relationships.
Chetan Jinadatha indicated no relevant financial relationships.
Phi Tran, DO1, Collin Telchik, MD1, Munok Hwang, MS2, Hosoon Choi, PhD2, Piyali Chatterjee, PhD2, Philip Toltzis, MD3, Curtis Donskey, MD4, Jennifer Cadnum, MS3, Christine Marlow, MS4, Chetan Jinadatha, MD2. P0202 - Antimicrobial Resistance in <i>Clostridioides difficile</i>: Is It on the Rise?, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
