Sunday Poster Session
Category: Stomach
P1601 - Development and External Validation of a Machine Learning-Based Gastric Cancer Prediction Model using Electronic Health Record Data
Sunday, October 27, 2024
3:30 PM - 7:00 PM ET
Location: Exhibit Hall E
Has Audio

Sarah Wehbe, MD
Cleveland Clinic
Cleveland, OH
Presenting Author(s)
Award: ACG Outstanding Research Award in the Stomach Category (Trainee)
Award: Presidential Poster Award
Sarah Wehbe, MD1, Sayf Al-deen Said, MD, MPH1, Carol Rouphael, MD2, John McMichael, MS1, Michelle Kang Kim, MD, PhD1
1Cleveland Clinic, Cleveland, OH; 2Digestive Disease Institute, Cleveland Clinic, Cleveland, OH
Introduction: In the United States, gastric cancer (GC) is associated with poor prognosis despite being curable if detected early. Population screening of GC could be feasible if there were a method to efficiently and accurately identify a high-risk cohort who would be eligible for GC screening. Such a model would need to demonstrate robust performance and be generalizable across the US population. In this study, we used data from the electronic health record (EHR) to develop and externally validate a machine learning-based risk prediction model for GC detection in a general adult population.
Methods: Non-cardia GC cases and controls ages 40 to 80 with at least one encounter at our medical centers in Ohio and Florida between 2010 and 2021 were identified using ICD 9/10 codes. After performing univariate analyses, we used data from the Ohio cohort to train and test multiple predictive models (logistic regression [LR], gradient boost [GB], random forest [RF], support vector [SV] and deep neural network [DNN]). Area Under the Receiver Operating Characteristic Curve (AUC ROC) was assessed for all models, and the best performing model was then externally validated using data from the Florida cohort.
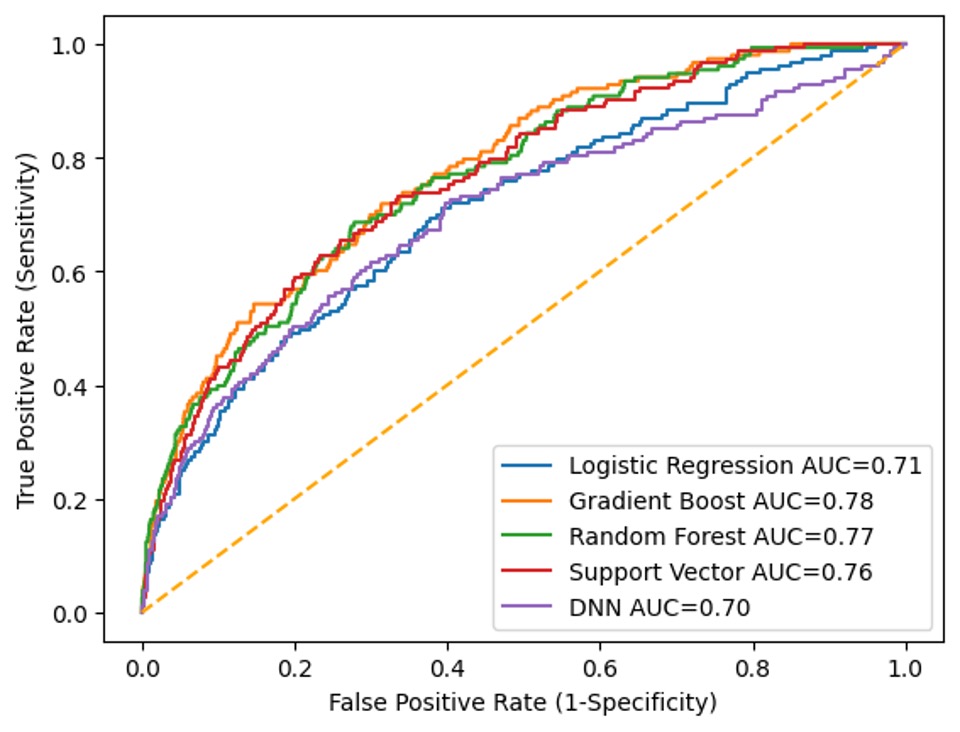
Results: 11,041 patients were included in the development set, of which 567 had GC and 10,474 were controls (Table). 2,716 patients were included in the validation set, of which 90 had GC and 2,626 were controls. In both cohorts, significant association was found between GC and the variables age, sex, race, ethnicity, body mass index and family history of GC. Helicobacter pylori was present in approximately 1% of the entire cohort. The GB model resulted in the best performance metrics on the development set with an AUC ROC of 0.78 (Figure). LR, RF, SV and DNN, achieved an AUC ROC of 0.71, 0.77, 0.76 and 0.70, respectively. In the external validation set, the GB model achieved a similar AUC ROC of 0.78.
Discussion: We demonstrate the robust performance and generalizability of a machine-learning based model for GC risk prediction using variables readily available in the EHR. Despite the demographic differences between the Ohio and Florida cohorts, the model exhibited solid performance in the external validation. Future studies will be performed to further refine this model. Ultimately, this risk prediction model may identify high-risk individuals who would be eligible for GC screening.
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Sarah Wehbe, MD1, Sayf Al-deen Said, MD, MPH1, Carol Rouphael, MD2, John McMichael, MS1, Michelle Kang Kim, MD, PhD1. P1601 - Development and External Validation of a Machine Learning-Based Gastric Cancer Prediction Model using Electronic Health Record Data, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
Award: Presidential Poster Award
Sarah Wehbe, MD1, Sayf Al-deen Said, MD, MPH1, Carol Rouphael, MD2, John McMichael, MS1, Michelle Kang Kim, MD, PhD1
1Cleveland Clinic, Cleveland, OH; 2Digestive Disease Institute, Cleveland Clinic, Cleveland, OH
Introduction: In the United States, gastric cancer (GC) is associated with poor prognosis despite being curable if detected early. Population screening of GC could be feasible if there were a method to efficiently and accurately identify a high-risk cohort who would be eligible for GC screening. Such a model would need to demonstrate robust performance and be generalizable across the US population. In this study, we used data from the electronic health record (EHR) to develop and externally validate a machine learning-based risk prediction model for GC detection in a general adult population.
Methods: Non-cardia GC cases and controls ages 40 to 80 with at least one encounter at our medical centers in Ohio and Florida between 2010 and 2021 were identified using ICD 9/10 codes. After performing univariate analyses, we used data from the Ohio cohort to train and test multiple predictive models (logistic regression [LR], gradient boost [GB], random forest [RF], support vector [SV] and deep neural network [DNN]). Area Under the Receiver Operating Characteristic Curve (AUC ROC) was assessed for all models, and the best performing model was then externally validated using data from the Florida cohort.
Results: 11,041 patients were included in the development set, of which 567 had GC and 10,474 were controls (Table). 2,716 patients were included in the validation set, of which 90 had GC and 2,626 were controls. In both cohorts, significant association was found between GC and the variables age, sex, race, ethnicity, body mass index and family history of GC. Helicobacter pylori was present in approximately 1% of the entire cohort. The GB model resulted in the best performance metrics on the development set with an AUC ROC of 0.78 (Figure). LR, RF, SV and DNN, achieved an AUC ROC of 0.71, 0.77, 0.76 and 0.70, respectively. In the external validation set, the GB model achieved a similar AUC ROC of 0.78.
Discussion: We demonstrate the robust performance and generalizability of a machine-learning based model for GC risk prediction using variables readily available in the EHR. Despite the demographic differences between the Ohio and Florida cohorts, the model exhibited solid performance in the external validation. Future studies will be performed to further refine this model. Ultimately, this risk prediction model may identify high-risk individuals who would be eligible for GC screening.

Figure: Performance Metrics of Multiple Predictive Models on the Development Set
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Sarah Wehbe indicated no relevant financial relationships.
Sayf Al-deen Said indicated no relevant financial relationships.
Carol Rouphael indicated no relevant financial relationships.
John McMichael indicated no relevant financial relationships.
Michelle Kang Kim indicated no relevant financial relationships.
Sarah Wehbe, MD1, Sayf Al-deen Said, MD, MPH1, Carol Rouphael, MD2, John McMichael, MS1, Michelle Kang Kim, MD, PhD1. P1601 - Development and External Validation of a Machine Learning-Based Gastric Cancer Prediction Model using Electronic Health Record Data, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.

