Tuesday Poster Session
Category: Colon
P3636 - Clustering Patients With Colonic Adenocarcinoma Using Unsupervised Machine Learning and Its Effect on Prognosis: A Population-Based Study
Tuesday, October 29, 2024
10:30 AM - 4:00 PM ET
Location: Exhibit Hall E

Has Audio
- ME
Marawan Elmassry, MBBCh
Texas Tech University Health Sciences Center
Lubbock, TX
Presenting Author(s)
Mohamed Mortagy, MBBCh1, Marawan Elmassry, MBBCh2, John Ramage, MBBS, PhD3
1Hampshire Hospitals NHS Foundation Trust, Winchester, England, United Kingdom; 2Texas Tech University Health Sciences Center, Lubbock, TX; 3Hampshire Hospitals NHS Foundation Trust, Basingstoke, England, United Kingdom
Introduction: Colonic Adenocarcinoma (CA) has an annual incidence of 1.7 per 100,000 individuals in the USA. The current AJCC UICC 8th edition TNM staging system doesn’t take grade or tumor size into account. K-means clustering is an unsupervised machine learning technique that helps identify patterns in patient populations leading to novel subgroups or staging of patients.
Methods: A total of 313,356 adult patients diagnosed with CA from 2000 to 2021 were extracted from the U.S. Surveillance, Epidemiology, and End Results (SEER) database registries after excluding those with missing data. These patients were clustered into two groups (0, 1) using K-means clustering. Clustering was evaluated using silhouette, Davies-Bouldin, Calinski-Harabasz scores and was visualized using principal Component Analysis (PCA) and T-distributed Stochastic Neighbor Embedding (T-SNE). SHapley Additive exPlanations (SHAP) showed the most important clinical features for clustering. Sixty-month survival was calculated for each cluster using Cox regression and Kaplan-Meier (KM) estimates. The impact of chemotherapy and surgery on each cluster was visualized with Kaplan-Meier curves. Using supervised machine learning, a decision tree was generated to categorize patients into one of the 2 clusters.
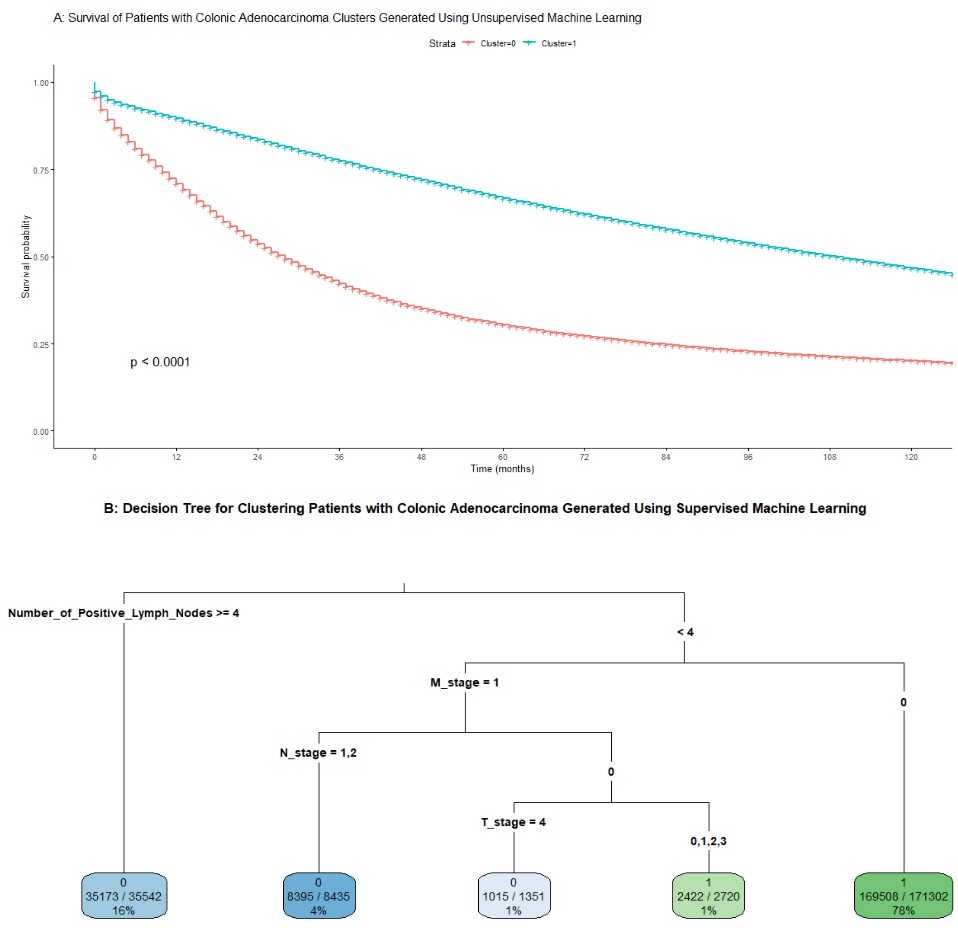
Results: The cohort characteristics and details for each cluster are shown in Table 1. PCA and T-SNE visualizations showed distinct clusters with some overlap. SHAP analysis indicated that the most important variables for clustering were N stage, number of positive lymph nodes, T stage, M stage, grade, size, age and presence of liver metastases in order. Kaplan Meier graphs showed statistically significant differences between the two clusters (Figure 1). The highest 60-month survival rate was observed in Cluster 1 (66.8%), followed by Cluster 0 (30.4%). The decision tree showed balanced accuracy of 97.5% in clustering patients (Figure 1). Patients receiving surgery (subtotal or total colectomy), or chemotherapy demonstrated improved survival in both clusters. This study is limited by some missing and/or inaccurate variables in the SEER database (e.g., treatment variables, co-morbidities) and its retrospective nature.
Discussion: K-means clustering of CA patients could be used in creating a novel staging system, predicting survival outcomes, and evaluating treatment effects within patient clusters.
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Mohamed Mortagy, MBBCh1, Marawan Elmassry, MBBCh2, John Ramage, MBBS, PhD3. P3636 - Clustering Patients With Colonic Adenocarcinoma Using Unsupervised Machine Learning and Its Effect on Prognosis: A Population-Based Study, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
1Hampshire Hospitals NHS Foundation Trust, Winchester, England, United Kingdom; 2Texas Tech University Health Sciences Center, Lubbock, TX; 3Hampshire Hospitals NHS Foundation Trust, Basingstoke, England, United Kingdom
Introduction: Colonic Adenocarcinoma (CA) has an annual incidence of 1.7 per 100,000 individuals in the USA. The current AJCC UICC 8th edition TNM staging system doesn’t take grade or tumor size into account. K-means clustering is an unsupervised machine learning technique that helps identify patterns in patient populations leading to novel subgroups or staging of patients.
Methods: A total of 313,356 adult patients diagnosed with CA from 2000 to 2021 were extracted from the U.S. Surveillance, Epidemiology, and End Results (SEER) database registries after excluding those with missing data. These patients were clustered into two groups (0, 1) using K-means clustering. Clustering was evaluated using silhouette, Davies-Bouldin, Calinski-Harabasz scores and was visualized using principal Component Analysis (PCA) and T-distributed Stochastic Neighbor Embedding (T-SNE). SHapley Additive exPlanations (SHAP) showed the most important clinical features for clustering. Sixty-month survival was calculated for each cluster using Cox regression and Kaplan-Meier (KM) estimates. The impact of chemotherapy and surgery on each cluster was visualized with Kaplan-Meier curves. Using supervised machine learning, a decision tree was generated to categorize patients into one of the 2 clusters.
Results: The cohort characteristics and details for each cluster are shown in Table 1. PCA and T-SNE visualizations showed distinct clusters with some overlap. SHAP analysis indicated that the most important variables for clustering were N stage, number of positive lymph nodes, T stage, M stage, grade, size, age and presence of liver metastases in order. Kaplan Meier graphs showed statistically significant differences between the two clusters (Figure 1). The highest 60-month survival rate was observed in Cluster 1 (66.8%), followed by Cluster 0 (30.4%). The decision tree showed balanced accuracy of 97.5% in clustering patients (Figure 1). Patients receiving surgery (subtotal or total colectomy), or chemotherapy demonstrated improved survival in both clusters. This study is limited by some missing and/or inaccurate variables in the SEER database (e.g., treatment variables, co-morbidities) and its retrospective nature.
Discussion: K-means clustering of CA patients could be used in creating a novel staging system, predicting survival outcomes, and evaluating treatment effects within patient clusters.

Figure: Figure 1 -
A: Kaplan Meier graph for survival of colon adenocarcinoma clusters generated using unsupervised machine learning
B:Decision tree generated using supervised machine learning to cluster patients with colonic adenocarcinoma
A: Kaplan Meier graph for survival of colon adenocarcinoma clusters generated using unsupervised machine learning
B:Decision tree generated using supervised machine learning to cluster patients with colonic adenocarcinoma
Note: The table for this abstract can be viewed in the ePoster Gallery section of the ACG 2024 ePoster Site or in The American Journal of Gastroenterology's abstract supplement issue, both of which will be available starting October 27, 2024.
Disclosures:
Mohamed Mortagy indicated no relevant financial relationships.
Marawan Elmassry indicated no relevant financial relationships.
John Ramage indicated no relevant financial relationships.
Mohamed Mortagy, MBBCh1, Marawan Elmassry, MBBCh2, John Ramage, MBBS, PhD3. P3636 - Clustering Patients With Colonic Adenocarcinoma Using Unsupervised Machine Learning and Its Effect on Prognosis: A Population-Based Study, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
