Tuesday Poster Session
Category: Colorectal Cancer Prevention
P3848 - AI-Assisted Survival Analysis on CRC gene sets From the TCGA Database
Tuesday, October 29, 2024
10:30 AM - 4:00 PM ET
Location: Exhibit Hall E
Has Audio

Jaydeep Mahasamudram, MD
BronxCare Health System
Rochester, MN
Presenting Author(s)
Jaydeep Mahasamudram, MD1, Manasvi Sharma, 2, Subhalakshmi Kandavel, MSc2, Manish Mishra, 2
1BronxCare Health System, Rochester, MN; 2Aganitha Cognitive Solutions, Hyderabad, Telangana, India
Introduction: Colorectal cancer (CRC) is a leading cause of cancer mortality worldwide, with a low survival rate for late-stage diagnoses. Identifying robust prognostic biomarkers is crucial for improving outcomes. The Cancer Genome Atlas (TCGA) project has generated comprehensive multi-omics data for CRC, allowing novel insights into its molecular landscape. AI techniques offer powerful approaches for analyzing large-scale omics data. In this study, we leveraged the TCGA CRC dataset and AI tools to perform survival analysis on gene expression profiles, aiming to identify robust prognostic gene signatures and elucidate their roles in CRC progression and therapeutic response.
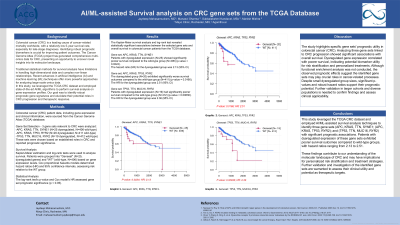
Methods: CRC patient data were sourced from TCGA. 3 gene sets relevant to CRC were analyzed: APC, KRAS, TTN, SYNE1; APC, KRAS, TP53, RYR2; TP53, TTN, MUC16, RYR2. Kaplan-Meier estimation, log-rank tests, and Cox proportional hazards models were used for survival analysis. Patients were grouped into "Geneset" (dysregulated genes) and "WT" (wild-type). Log-rank p-value and Cox HR assessed prognostic significance (p < 0.05).
Results: Kaplan-Meier and log-rank tests revealed significant associations between the gene sets and overall survival.
APC, KRAS, TTN, SYNE1: Dysregulated group had poorer survival vs WT (p=0.0224). HR=2.13, 2.13-fold increased risk.
APC, KRAS, TP53, RYR2: Dysregulated group had worse survival vs WT (p=0.0166). HR=2.51, 2.51-fold higher risk.
TP53, TTN, MUC16, RYR2: Dysregulated group had poorer survival vs WT (p=0.00836). HR=2.36, 2.36-fold increased risk.
Discussion: The study highlights specific gene sets' prognostic utility in CRC. Dysregulated expression correlated with poorer survival, indicating potential biomarker utility for risk stratification and personalized treatments. The observed prognostic effects suggest the identified gene sets may play crucial roles in cancer-related processes. Despite small dysregulated group sizes, significant p-values and robust hazard ratios support their prognostic potential. Further validation is needed to assess clinical applicability.

Disclosures:
Jaydeep Mahasamudram, MD1, Manasvi Sharma, 2, Subhalakshmi Kandavel, MSc2, Manish Mishra, 2. P3848 - AI-Assisted Survival Analysis on CRC gene sets From the TCGA Database, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
1BronxCare Health System, Rochester, MN; 2Aganitha Cognitive Solutions, Hyderabad, Telangana, India
Introduction: Colorectal cancer (CRC) is a leading cause of cancer mortality worldwide, with a low survival rate for late-stage diagnoses. Identifying robust prognostic biomarkers is crucial for improving outcomes. The Cancer Genome Atlas (TCGA) project has generated comprehensive multi-omics data for CRC, allowing novel insights into its molecular landscape. AI techniques offer powerful approaches for analyzing large-scale omics data. In this study, we leveraged the TCGA CRC dataset and AI tools to perform survival analysis on gene expression profiles, aiming to identify robust prognostic gene signatures and elucidate their roles in CRC progression and therapeutic response.
Methods: CRC patient data were sourced from TCGA. 3 gene sets relevant to CRC were analyzed: APC, KRAS, TTN, SYNE1; APC, KRAS, TP53, RYR2; TP53, TTN, MUC16, RYR2. Kaplan-Meier estimation, log-rank tests, and Cox proportional hazards models were used for survival analysis. Patients were grouped into "Geneset" (dysregulated genes) and "WT" (wild-type). Log-rank p-value and Cox HR assessed prognostic significance (p < 0.05).
Results: Kaplan-Meier and log-rank tests revealed significant associations between the gene sets and overall survival.
APC, KRAS, TTN, SYNE1: Dysregulated group had poorer survival vs WT (p=0.0224). HR=2.13, 2.13-fold increased risk.
APC, KRAS, TP53, RYR2: Dysregulated group had worse survival vs WT (p=0.0166). HR=2.51, 2.51-fold higher risk.
TP53, TTN, MUC16, RYR2: Dysregulated group had poorer survival vs WT (p=0.00836). HR=2.36, 2.36-fold increased risk.
Discussion: The study highlights specific gene sets' prognostic utility in CRC. Dysregulated expression correlated with poorer survival, indicating potential biomarker utility for risk stratification and personalized treatments. The observed prognostic effects suggest the identified gene sets may play crucial roles in cancer-related processes. Despite small dysregulated group sizes, significant p-values and robust hazard ratios support their prognostic potential. Further validation is needed to assess clinical applicability.

Figure: Figure A : Geneset: APC, KRAS, TTN, SYNE1
Figure B : Geneset: APC, KRAS, TP53, RYR2
Figure C : Geneset: TP53, TTN, MUC16, RYR2
Figure B : Geneset: APC, KRAS, TP53, RYR2
Figure C : Geneset: TP53, TTN, MUC16, RYR2
Disclosures:
Jaydeep Mahasamudram indicated no relevant financial relationships.
Manasvi Sharma indicated no relevant financial relationships.
Subhalakshmi Kandavel indicated no relevant financial relationships.
Manish Mishra indicated no relevant financial relationships.
Jaydeep Mahasamudram, MD1, Manasvi Sharma, 2, Subhalakshmi Kandavel, MSc2, Manish Mishra, 2. P3848 - AI-Assisted Survival Analysis on CRC gene sets From the TCGA Database, ACG 2024 Annual Scientific Meeting Abstracts. Philadelphia, PA: American College of Gastroenterology.
